L2E convex regression
L2E_convex.RdL2E_convex performs convex regression under the L2 criterion. Available methods include proximal gradient descent (PG) and majorization-minimization (MM).
L2E_convex( y, beta, tau, method = "MM", max_iter = 100, tol = 1e-04, Show.Time = TRUE )
Arguments
| y | Response vector |
|---|---|
| beta | Initial vector of regression coefficients |
| tau | Initial precision estimate |
| method | Available methods include PG and MM. MM by default. |
| max_iter | Maximum number of iterations |
| tol | Relative tolerance |
| Show.Time | Report the computing time |
Value
Returns a list object containing the estimates for beta (vector) and tau (scalar), the number of outer block descent iterations until convergence (scalar), and the number of inner iterations per outer iteration for updating beta (vector) and tau or eta (vector)
Examples
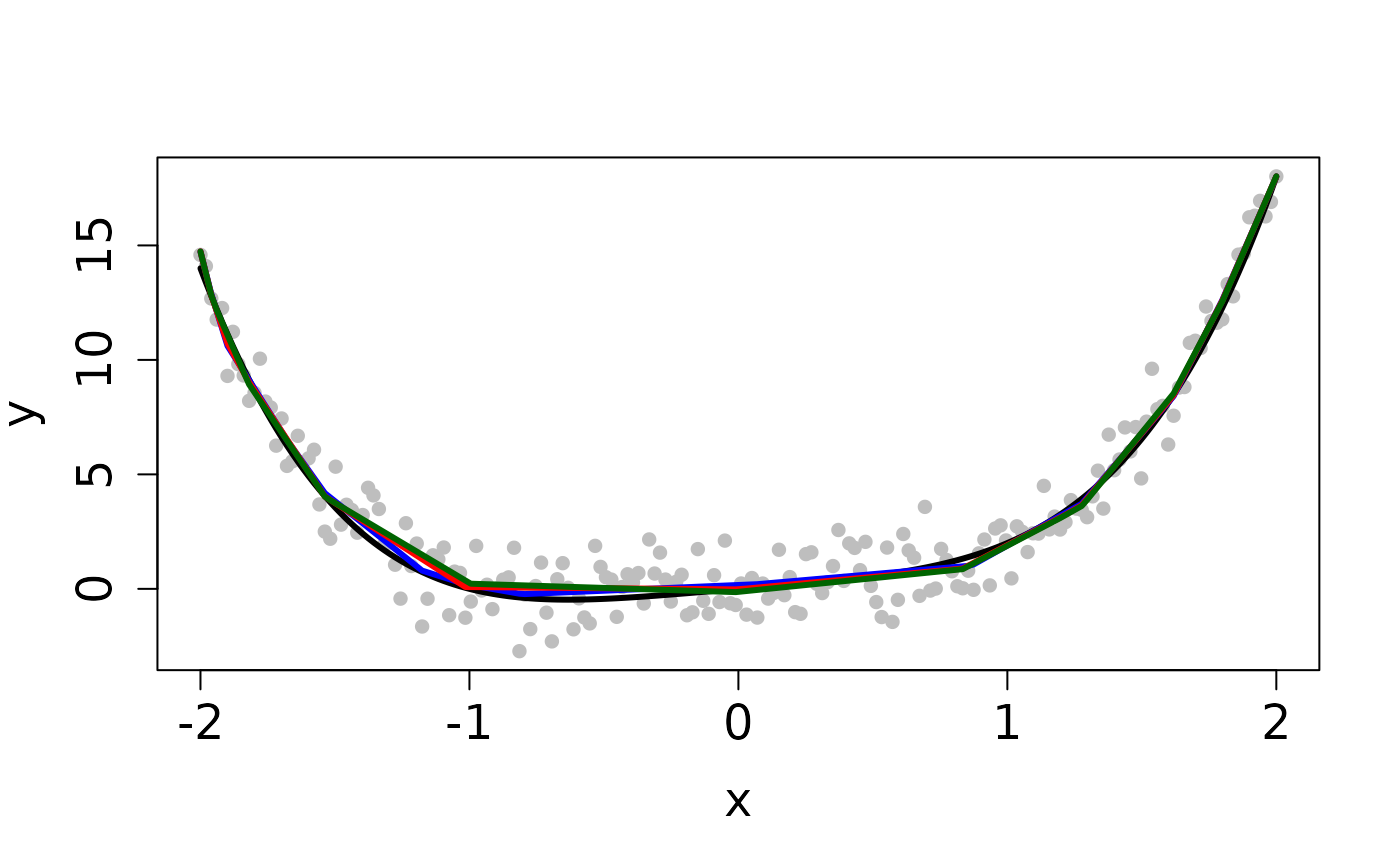
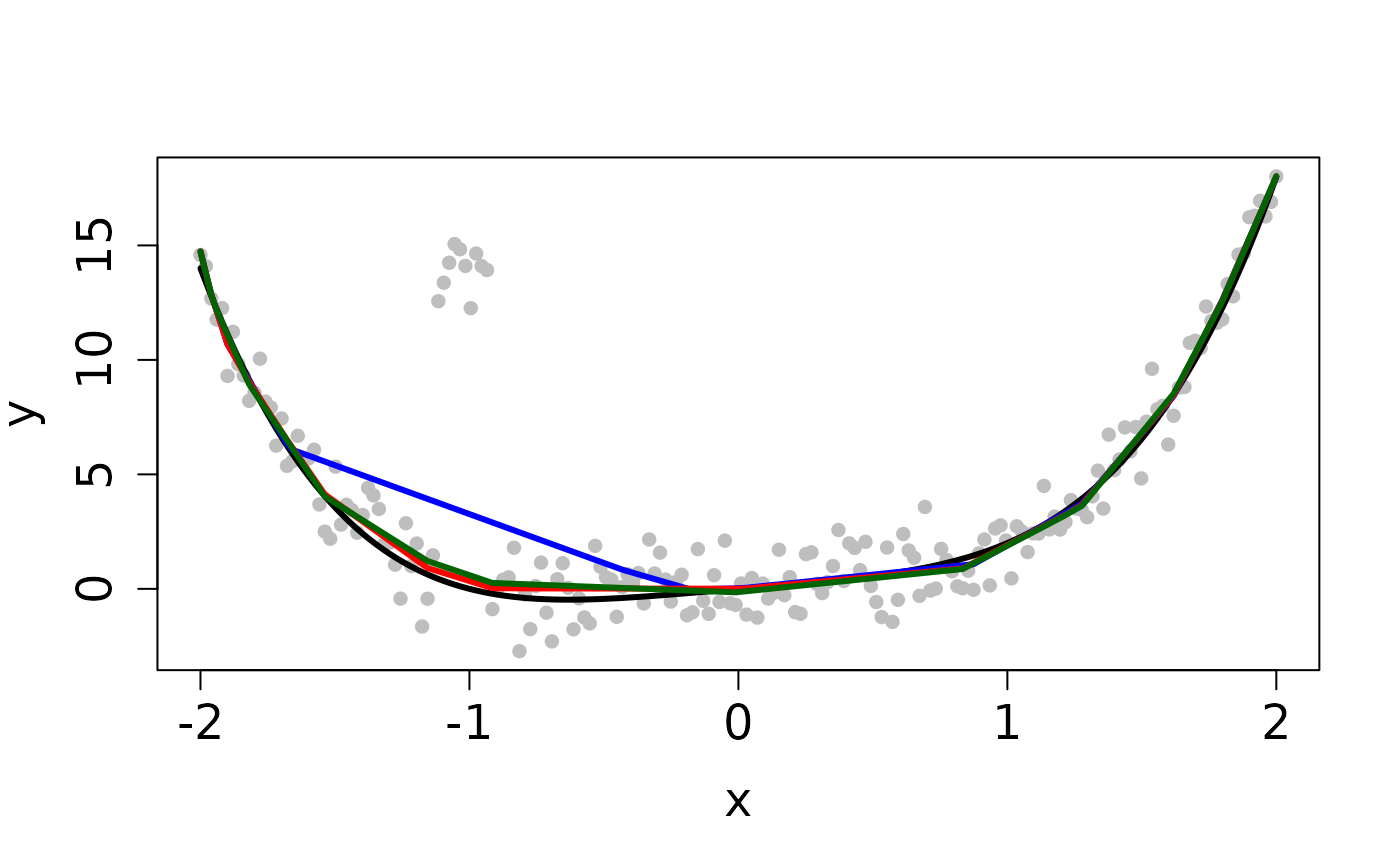
set.seed(12345) n <- 200 tau <- 1 x <- seq(-2, 2, length.out=n) f <- x^4 + x y <- f + (1/tau) * rnorm(n) ## Clean Data plot(x, y, pch=16, cex.lab=1.5, cex.axis=1.5, cex.sub=1.5, col='gray')tau <- 1 b <- y ## Least Squares method cvx <- fitted(cobs::conreg(y, convex=TRUE)) ## MM method sol_mm <- L2E_convex(y, b, tau)#> user system elapsed #> 0.057 0.000 0.056## PG method sol_pg <- L2E_convex(y, b, tau, method='PG')#> user system elapsed #> 0.155 0.000 0.156## Contaminated Data ix <- 0:9 y[45 + ix] <- 14 + rnorm(10) plot(x, y, pch=16, cex.lab=1.5, cex.axis=1.5, cex.sub=1.5, col='gray')#> user system elapsed #> 0.132 0.000 0.133sol_pg <- L2E_convex(y, b, tau, method='PG')#> user system elapsed #> 0.950 0.000 0.951